User Guide
Preparation of input file
We now support CSV, TXT, and MGF data types.
CSV data type appears as follows:
192,0.02
193,0.15
194,14.38
195,1.89
196,0.19
TXT data type appears as follows:
107.0 0.0034
108.0 0.0015
109.0 0.0288
110.0 1.0
111.0 0.0726
112.0 0.0068
MGF data type should contain at least the COMPOUND_NAME attribute.
BEGIN IONS
COMPOUND_NAME=substance_A
108.0 0.0015
109.0 0.0288
110.0 1.0
111.0 0.0726
112.0 0.0068
END IONS
BEGIN IONS
COMPOUND_NAME=substance_B
213.0 0.0013
215.0 0.0382
216.0 1.0
217.0 0.1182
218.0 0.9887
219.0 0.0896
220.0 0.0073
END IONS
Example data can be found here: Example Data
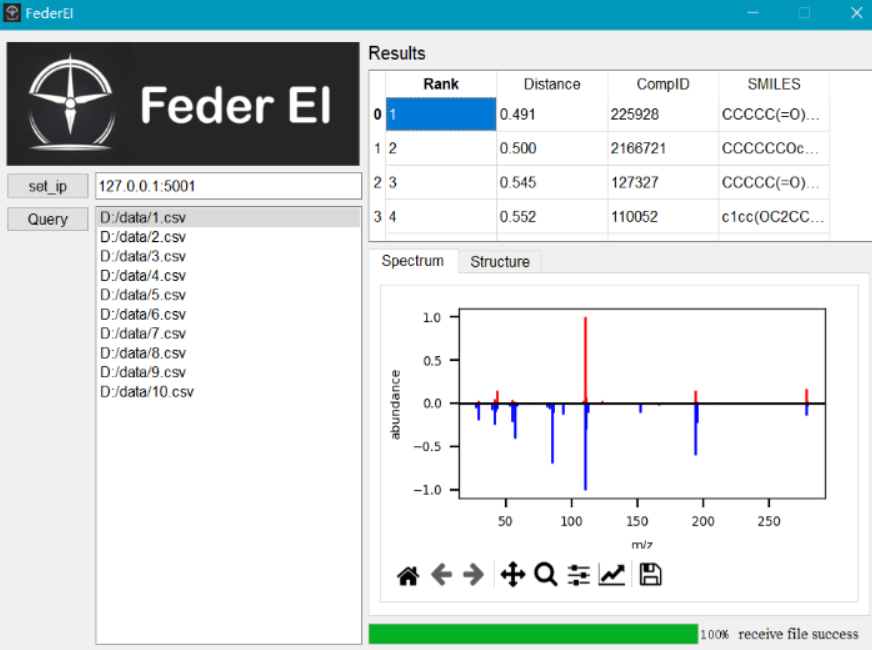
Operation sequence
- First, set the central IP and port.
- Second, input query data.
- Third, wait and view the result.
Interpretation of results
Clicking the query data will display the results. Clicking on a result will show a comparison of mass spectra. The compound structure will also be displayed.
Operation demonstration
Saving results
Result files will automatically be saved in the same folder as the FederEI.exe file.
The filename consists of a random number and the query time, designed to prevent overlapping results with the same name.
The file content includes the query results for all compounds. This .mgf file contains the following information:
smiles
compound_name
distance
origin_name
ms
SMILES, compound_name, and MS come from the corresponding compounds in the Library, while "distance" indicates the gap between the queried compound and the corresponding compound in the Library. "origin_name" is the name of the queried compound. If .csv or .txt file is used for the query, it corresponds to the filename; if an .mgf file is used, it corresponds to the original compound name.